The team led by Prof. Chenlong Li from School of Life Sciences made new progress on how the Polycomb-mediated gene silencing is established in plants
Source: School of Life Sciences
Edited by: Tan Rongyu, Wang Dongmei
Polycomb group (PcG) proteins constitute a global and evolutionarily conserved transcriptional repression system that is essential for growth and development in multicellular organisms ranging from plants to humans. PcG proteins assemble into several types of multi-protein chromatin-modifying complexes to confer repressive histone modifications and compact chromatin at target loci. The two main types of Polycomb complexes are Polycomb repressive complex 1 (PRC1) and PRC2. PRC2 catalyzes trimethylation at H3 lysine 27 (H3K27me3), whereas PRC1 catalyzes H2A lysine 119 ubiquitination. The two complexes are intimately linked, and their combined activities are essential for Polycomb-mediated gene silencing. The failure of the establishment and/or maintenance of H3K27me3 at Polycomb target loci leads to derepression of the transcriptional activity of genes at developmental stages when they should be silenced. Despite the importance and conservation of the Polycomb machinery in gene silencing and cellular differentiation, how it recognizes its target loci is still not fully understood.
Recently, the team led by Prof. Chenlong Li from the School of Life Sciences, Sun Yat-sen University, reveal important mechanistic insights into the establishment of Polycomb silencing in plants. They report genome-wide evidence for the recruitment of PRC2 by the transcriptional repressors VIVIPAROUS1/ABI3-LIKE1 (VAL1) and VAL2 in
Arabidopsis thaliana, a model flowering plant. They show that the
val1 val2 double mutant possesses somatic embryonic phenotypes and a transcriptome strikingly similar to those of the
swn clf double mutant, which lacks the PRC2 catalytic subunits SWINGER (SWN) and CURLY LEAF (CLF). They further show that VAL1 and VAL2 physically interact with SWN and CLF in vivo (Figure 1). Genome-wide binding profiling demonstrated that they colocalize with SWN and CLF at PRC2 target loci (Figure 1). Loss of VAL1/2 significantly reduces SWN and CLF enrichment at PRC2 target loci and leads to a genome-wide redistribution of H3K27me3 that strongly affects transcription. Finally, they provide evidence that the VAL1/VAL2–RY regulatory system is largely independent of previously identified modules for Polycomb silencing in plants. Together, their work uncovered the mechanistic basis of VAL function in mediating Polycomb silencing at a genome-wide scale that would be important for understanding how plant genes can be silenced by PcG proteins during development (Figure 2).
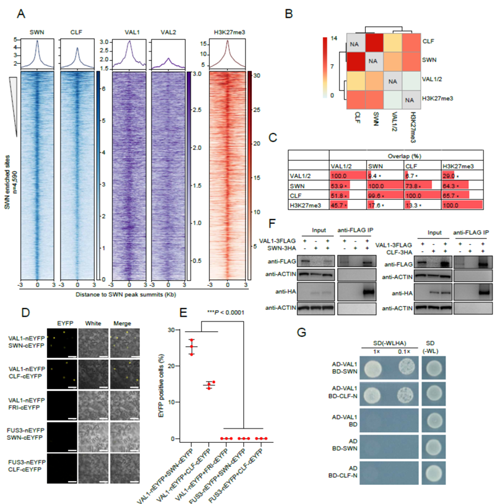
Figure 1: The VAL1/2 directly interact with PRC2 catalytic subunits SWN and CLF, and occupy a number of PRC2 target genes in Arabidopsis genome.
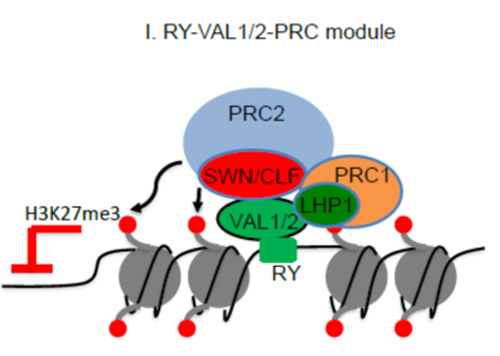
Figure 2: The working model showing how VAL1/2 recruit PRC2 complexes.
The work entitled“The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in
Arabidopsis”was published in
Nucleic Acids Research (IF:11.5) on Dec 4, 2020. Prof. Chenlong Li is the corresponding author. Dr. Liangbing Yuan is the first author. This work was supported by the National Natural Science Foundation of China, Guangdong Basic and Applied Basic Research Foundation, and Fundamental Research Funds for the Central Universities.
Link to the paper:
https://doi.org/10.1093/nar/gkaa1129


