Prof. Shi Xiao Research Group in School of Life Sciences Made New Progress in the Regulation Mechanism of Plant Autophagy
Source: School of Life Sciences
Written by: School of Life Sciences
Edited by: Wang Dongmei
Recently, a research article entitled “Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13” has been published online in The Plant Cell, one of the top journals in plant field. This study accomplished by Prof. Shi Xiao from School of Life Sciences at Sun Yat-sen University, revealed that the E3 ligases SINAT family proteins regulate autophagy initiation and termination dynamics by modulating the ATG1-ATG13 kinase complex ubiquitylation and degradation. This research elucidated the regulation mechanism of ATG1-ATG13 homeostasis, providing an important theoretical basis for understanding the molecular mechanism of autophagy in plants.
Autophagy is a key biological process to maintain eukaryotic cell homeostasis, which plays an important role in regulating plant growth, development, and in response to biotic and abiotic stresses. Nevertheless, little is known about the regulation mechanism of autophagy related proteins.
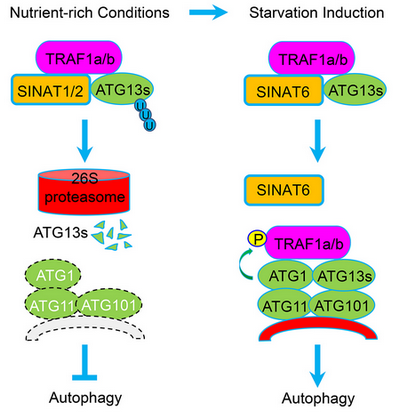
A Working Model Showing the SINAT proteins in the Regulation of Autophagy Dynamics in Arabidopsis
Through genetic and biochemical analysis, the team of Prof. Shi Xiao found that the stability of ATG1 and ATG13 proteins, members of the autophagy protein kinase complex, were dynamically regulated by the 26S proteasome pathway under both starvation conditions and during recovery after starvation treatments. Under normal growth conditions, the E3 ubiquitin ligases SINAT1 and SINAT2 promoted the degradation of ATG13 through K48-linked ubiquitylation by interacting with ATG13, and suppression of autophagy. Under starvation conditions, SINAT6 inhibited the ubiquitylation and degradation of ATG13 and induced autophagosome biogenesis. Further study revealed that two lysine residues of ATG13a protein, K607 and K609, were crucial for ATG13 ubiquitylation and inhibition of autophagy. Further, adaptors TRAF1a and TRAF1b, were involved in regulating the stability of ATG13 mediated by SINAT1/SINAT2 and SINAT6. Meanwhile, ATG1 protein kinase promoted TRAF1a protein stability by phosphorylation-like modification in vivo in a feedback mechanism under nutrient starvation conditions, further regulated the homeostasis of autophagy in plants (as shown in the figure above).
Consistent with ATGs functioning in autophagy, the Arabidopsis atg1a atg1b atg1c triple knockout mutants exhibited premature leaf senescence, hypersensitivity to nutrient starvation, and reduction in TRAF1a stability. Therefore, these findings demonstrate that SINAT family proteins facilitate ATG13 ubiquitylation and stability and thus regulate autophagy, which is of great scientific significance.
This work was supported by The National Science Fund for Distinguished Young Scholars Program, National Natural Science Foundation of China Program, National Natural Science Foundation of China Youth Program, and Three Major Construction Projects of Sun Yat-sen University. Postdoctor Hua Qi and visiting research fellow Juan Li are co-first authors, Prof. Shi Xiao is the corresponding author. Prof. Qingming Zhou from Hunan Agricultural University participates in part of this work.
